My Research
I study deep-time evolutionary biology, exploring the diversification of life on Earth by combining research on genomes and fossils.
My research encompasses phylogenomics, macroevolution, and paleobiology using cutting-edge computational, sequencing, and imaging techniques. Although much of my focus is on sea urchins and other echinoderms, I enjoy working across the invertebrate tree of life.
Scientific Path
Throughout my years as a scientist, I have been part of biology, geology, and oceanography departments, solidifying a interdisciplinary perspective that I strive to maintain as my scientific journey continues. This has also given me the opportunity to teach a wide range of courses on the topics of phylogenetics, evolution, paleobiology, statistics, and computer science.
Licenciatura (BS+MS)
Facultad de Ciencias Exactas y Naturales
Universidad de Buenos Aires
MPhil & PhD
Department of Earth & Planetary Sciences
Yale University
Postdoctoral Fellow & Lecturer
Scripps Institution of Oceanography
UC San Diego
Research Scientist
Department of Ecology & Evolutionary Biology
Princeton University
Assistant Professor (soon…)
Department of Biology
Colorado State University
Research Goals
Reconstructing events that occurred millions of years ago is a daunting task. Echoes of distant pasts can be found today in two main places: the genomes of living organisms, which retain historical signals that can help us assemble the tree of life; and the geological record, a unique window into past states of the biosphere. Accurately reconstructing the past, and the trajectory connecting it to present-day conditions, necessitates the integration of these two distinct sources of information.
I strive to advance the field of phylogenetic paleobiology, developing novel methods and computational tools that help blur the paleontological-neontological divide and showcase the benefits of tackling evolutionary questions using integrative approaches. In the process, I also seek to establish sea urchins as a model clade for macroevolutionary research.
Latest News
Registration opened for a new edition of the online course Morphological Phylogenetic
The online platform Transmitting Science is hosting the 8th edition of the course “Morphological Phylogenetics: Principles, Applications, and Techniques” which will run for three weeks in March 2026. Register here to catch up with the exciting state of the field of morphological phylogenetics, covering everything from character construction to tip dating under the FBD model, and combining theory with hand-on exercises that introduce available software.
Participation in the palaeoverse Seminar Series
Thrilled to have given an invited online talk at the palaeoverse Seminar Series along with friend and colleague Russell J. Garwood (U of Manchester). Shut out to the organizers for bringing the paleontological community together around such great content and forward-thinking goals. For those interested in reconstructing not just the tree of life, but the tree of life and death, TREvoSim offers novel ways of simulating data to explore how diversification and fossilization occurred, and how much we can expect to succeed in reconstructing the past.
Began new position at Princeton EEB!
As of January 2025, I am a proud member of the Simões lab at Princeton University, working on morphological phylogenetics, genomes, and the fossil record. There is probably no better place to help advance the field of phylogenetic paleobiology.
The deep sea has three new stars * * *
Former MS student Zihui Shen (now doing a PhD at Hong Kong Baptist University) described three new sea stars from the genus Caymanostella, a poorly known deep sea clade including mostly wood associated species. One of them, Caymanostella scrippscogniaticausa, names after the Scripps family for their continued support to oceanographic science, was highlighted by the World Register of Marine Species as one of the 10 most remarkable new marine species of 2024.
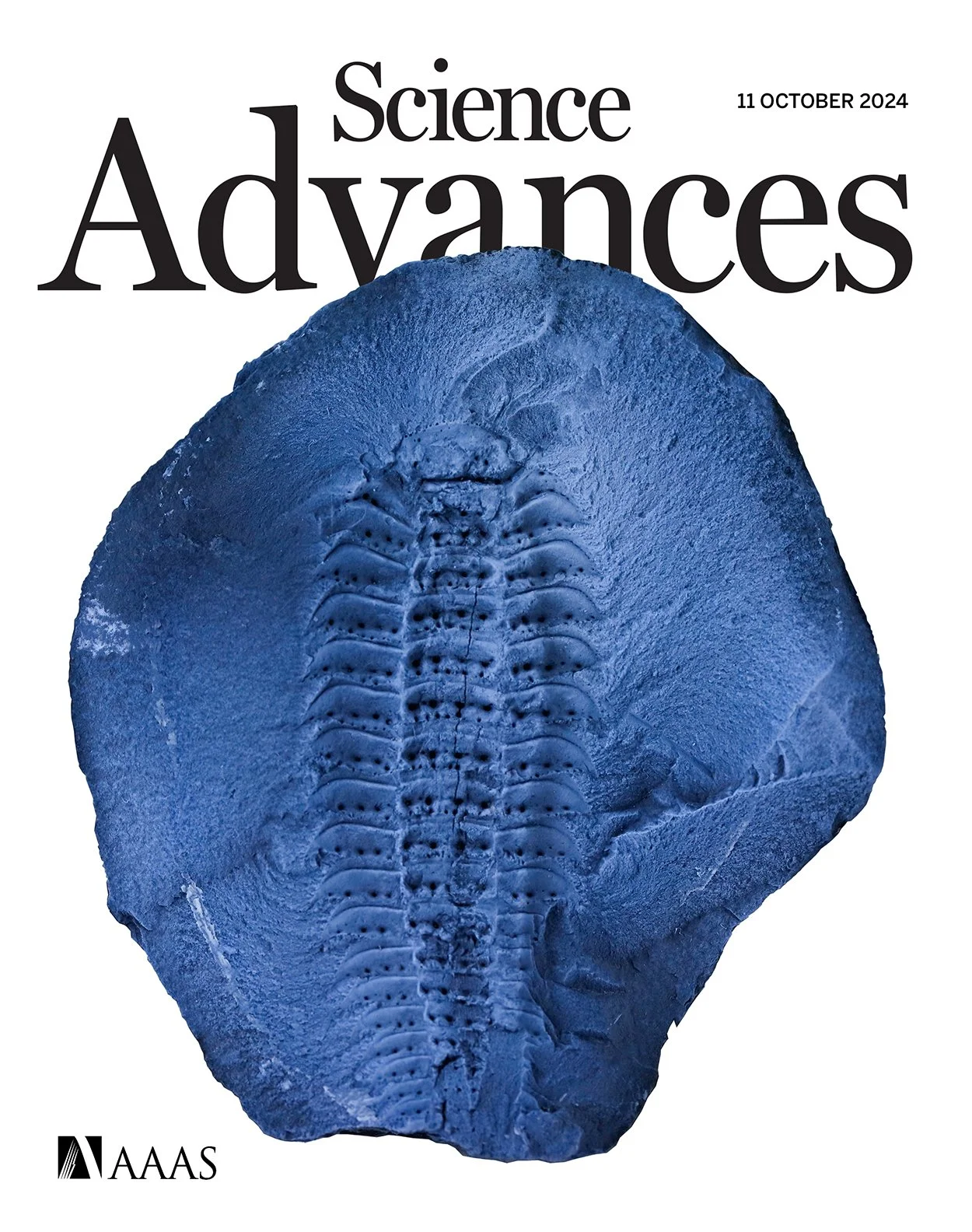
An exceptionally preserved giant myriapod from the Carboniferous and the diversification of centipedes and millipedes
Myriapod relationships have been notoriously hard to resolve, with strongly discordant results obtained using morphological and molecular efforts. Amazing fossil finds from Montceau-les-Mines (France), imaged using μCT technology, help trace the deep ancestry and morphological evolution of this unique group of terrestrial predators, highlighting the benefits of combining genomic and paleontological research. The work was highlighted on the cover of Scientific Advances! You can find a digest of the science here.
chronospace R package is out in Methods in Ecology & Evolution!
Divergence time estimation is a tricky thing. We have developed tools to help quantify and visualize the sensitivity of the method to methodological decisions, providing novel approaches of exploring uncertainty. Check the manuscript, download the package, and read more about it (and science in general) at this interview with the British Ecological Society who shortlisted the paper for the 2024 Robert May Prize.